Research: Aging Network Bioinformatics
The experiment
Previously I ran an experiment to find central genes from the OSK genes used to create induced pluripotent stem cells to the global aging genes (genes discovered in a paper by checking many aging tissues and seeing what genes are either up or down regulated in common across the different tissues). Now my question is, what are the genes central to the global aging genes? Does it link to the aging machinery that governs the cell?
So what we’ll do to find the central genes is compare the global aging genes to themselves in a pairwise fashion. To be more precise, it’ll be like having two lists of all the global aging genes and we’ll compare every pair between the two lists. When we compare the pairs, we’ll find the strongest path of interactions through the gene regulatory network. Then we aggregate these strongest paths by counting the nodes that appear over and over again.
The experiment is run on the global aging genes while the control is run on randomly selected genes from the regulatory network.
The data
The regulatory network is downloaded from https://grand.networkmedicine.org/tissues/ and I used the fibroblast cell line. Also I used the PANDA version and to download it under the “Network” column you click “Adj”.
The way I converted between ENSG ids and actual gene names I used https://ftp.ensembl.org/pub/release-74/gtf/homo_sapiens/. It uses an older genome assembly since that’s what the regulatory gene paths uses. Potentially could get better results if it all used a newer one. Since when I initially discovered the difference between how many genes mapped between assembly 28 to the gene regulatory pathways using assembly 27, there was like 25% that were not found.
I used the global aging gene data from this paper https://elifesciences.org/articles/62293 and the direct link to the data is here https://figshare.com/articles/dataset/tms_gene_data_rv1/12827615?file=27857814.
Finally I used the GenAge dataset that is a dataset of genes related to aging https://genomics.senescence.info/download.html.
The code
The code can be found here: https://github.com/luojxxx/Aging-machinery. The dependencies are all at the top. The specific ipython notebook is bioinfo_PANDA_GAG.ipynb
The code itself is relatively simple. It loads and cleans all the data. The only part that’s kind of interesting is how the regulatory networks are turned into something that works with the code to find the shortest path.
So the regulatory pathways comes to you as a table between the regulatory genes and it’s effect on all the other genes in the cell. Large numbers represent a large effect, small numbers a small effect; positive numbers represent a upregulating effect, negative numbers represent a downregulating effect. But how do we find the shortest path when you need the fewest nodes with the largest weights while some are upregulating and downregulating.
Basically you take the inverse of the numbers, so that the largest effects become the smallest number; this way you’re trying to get the fewest nodes with the smallest weights. Finally you take the absolute value of the weights, so that it’s just finding the strongest interactions whether it’s up or down regulating. Now you have a simple but properly represented graph you can plug into a shortest path algorithm.
Finally the control takes the regulatory pathway and samples randomly from the ~30,000 genes. There’s the same number of random genes as the global aging genes, so that it’s comparable. The control is run three times with different random genes.
If you want to run it yourself, just put the ipython notebook, ENSG dataset, and GAG dataset in the same folder, and then create a ‘data’ folder to store the Grand datasets.
The results
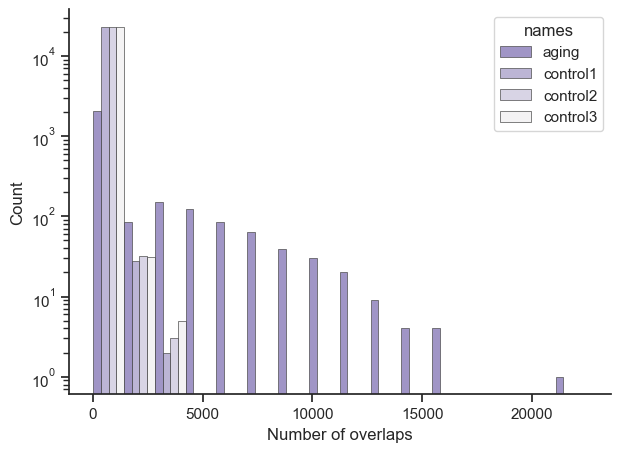
Overall, this is an exceptional result, central genes from the global aging genes are enriched/overlapped on average 10 times over the control sets.
Just so it’s clearer, let’s say you had a result of {‘ENSG1’: 10, ‘ENSG2’: 10, ‘ENSG3’: 50}. The X axis would show a bar at number of overlaps 10 with a count of 2, and another bar at number of overlaps 50 with count 1. Also the bars position is roughly accurate, due to putting multiple bars for aging, control1, control2, and control3 next to each other.
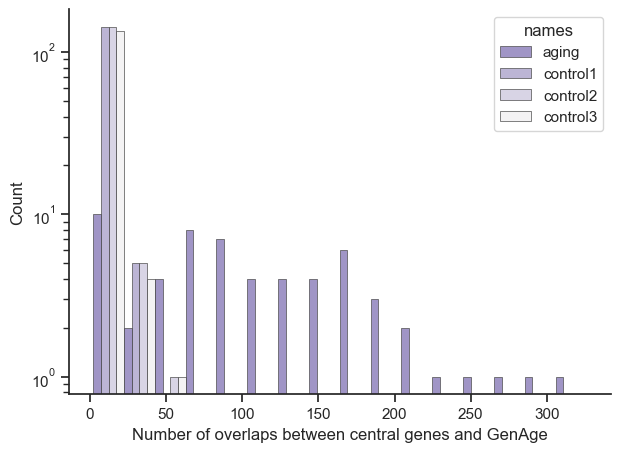
And to further drive home that aging related genes are being represented as central genes to the global aging genes, this is the comparison between the genes found in the GenAge dataset (a dataset of genes related to aging) and our results.
And again to be clear, let’s say you had a result where [‘ENSG1’: 10, ‘ENSG2’: 10, ‘ENSG3’: 20] and the GenAge dataset said [‘ENSG1’], then there’d be a bar at the 10 on the x-axis and a count of 1 on the y-axis.
Aging related genes are exceptionally enriched in the central genes from the global aging genes compared to the control set about ~20 times on average. This suggests that the results that aren’t already in the GenAge dataset could have potential aging related functions, providing a rich dataset for future exploration (in particular with this experiment).
RAW Output
For the sake of convenience, I’ve copied out the raw output from the jupyter notebook. You can just google search the ENSG ids to see what the gene does.
Global Aging Gene set
Size: 1599
Average: 62.79049405878674
Median: 6
Control set 1
Size: 14440
Average: 6.249445983379501
Median: 4.0
Control set 2
Size: 14396
Average: 6.286607390941929
Median: 4.0
Control set 3
Size: 14186
Average: 6.3752995911462005
Median: 4.0
Global Aging Gene set (filtered by GenAge)
Size: 59
Average: 113.15254237288136
Median: 98
Control set 1 (filtered by GenAge)
Size: 148
Average: 6.4324324324324325
Median: 4.0
Control set 2 (filtered by GenAge)
Size: 148
Average: 6.175675675675675
Median: 4.0
Control set 3 (filtered by GenAge)
Size: 139
Average: 6.733812949640288
Median: 4
Update (4/4/2025)
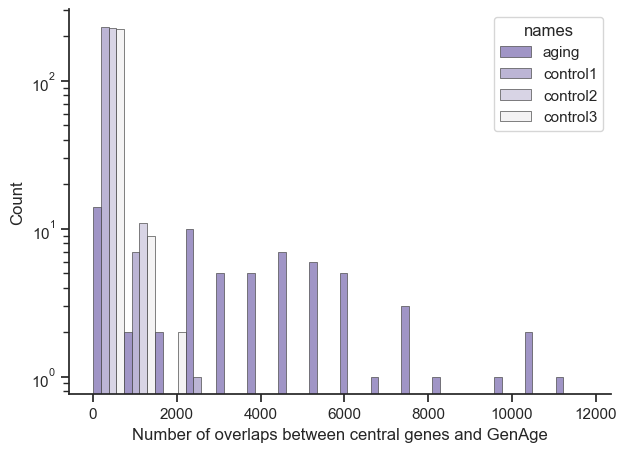
So after looking through the results I noticed that many of the genes found in the GenAge dataset that when mutated have diseases in specific organs. So to run this analysis only on a fibroblast cell line dataset might be too limited, so I re-ran the experiment on all 36 available tissues and the results are below. The specific code is bioinfo_PANDA_GAG_multiprocess_graphTool2.ipynb, and it uses a combination of a C backend network tool called graph_tool and multiprocessing to efficiently run through the analysis. The overall results are similar in terms of how enriched the global aging gene set is compared to the control set.
Just so it’s clearer, let’s say you had a result of {‘ENSG1’: 10, ‘ENSG2’: 10, ‘ENSG3’: 50}. The X axis would show a bar at number of overlaps 10 with a count of 2, and another bar at number of overlaps 50 with count 1. Also the bars position is roughly accurate, due to putting multiple bars for aging, control1, control2, and control3 next to each other.
And again to be clear, let’s say you had a result where [‘ENSG1’: 10, ‘ENSG2’: 10, ‘ENSG3’: 20] and the GenAge dataset said [‘ENSG1’], then there’d be a bar at the 10 on the x-axis and a count of 1 on the y-axis.
RAW Output
For the sake of convenience, I’ve copied out the raw output from the jupyter notebook. You can just google search the ENSG ids to see what the gene does.
Global Aging Gene set
Size: 2652
Average: 1368.1546003016592
Median: 20.0
Control set 1
Size: 23102
Average: 130.08423513115747
Median: 64.0
Control set 2
Size: 23068
Average: 131.1772151898734
Median: 64.0
Control set 3
Size: 22969
Average: 131.7830989594671
Median: 62
Global Aging Gene set (filtered by GenAge)
Size: 65
Average: 3849.7846153846153
Median: 3514
Control set 1 (filtered by GenAge)
Size: 241
Average: 159.38589211618256
Median: 66
Control set 2 (filtered by GenAge)
Size: 239
Average: 167.12970711297072
Median: 66
Control set 3 (filtered by GenAge)
Size: 237
Average: 171.17299578059072
Median: 74
Update (5/2/2025)
After thinking about it, I added one final metric to sift through the list of potential aging genes: pubmed search results. Basically I search for the gene name along with the keyword “aging” and count how many search results are returned. This number is divided by the maximum value found from across the whole dataset to return a percentile. The same is done for the overlaps, the overlap count for a particular gene is divided by the maximum value found across the whole dataset. The two percentiles are added together, each with a max value of 1, together with a max value of 2, and the results are sorted based on the combined hybrid value. The results are shown below along with a boolean that shows whether the gene is found in the GenAge dataset of identified genes related to aging. As you can see many of the top results are found in the GenAge dataset, which highlights how this enriching process works and how the yet unverified genes provides a shortlist for further research.